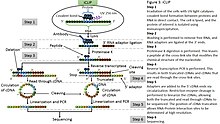
CLIP-seq overview. The CLIP-seq method for identifying miRNA target... | Download Scientific Diagram

Main steps of the bioinformatics workflow to analyze CLIP-seq data with... | Download Scientific Diagram

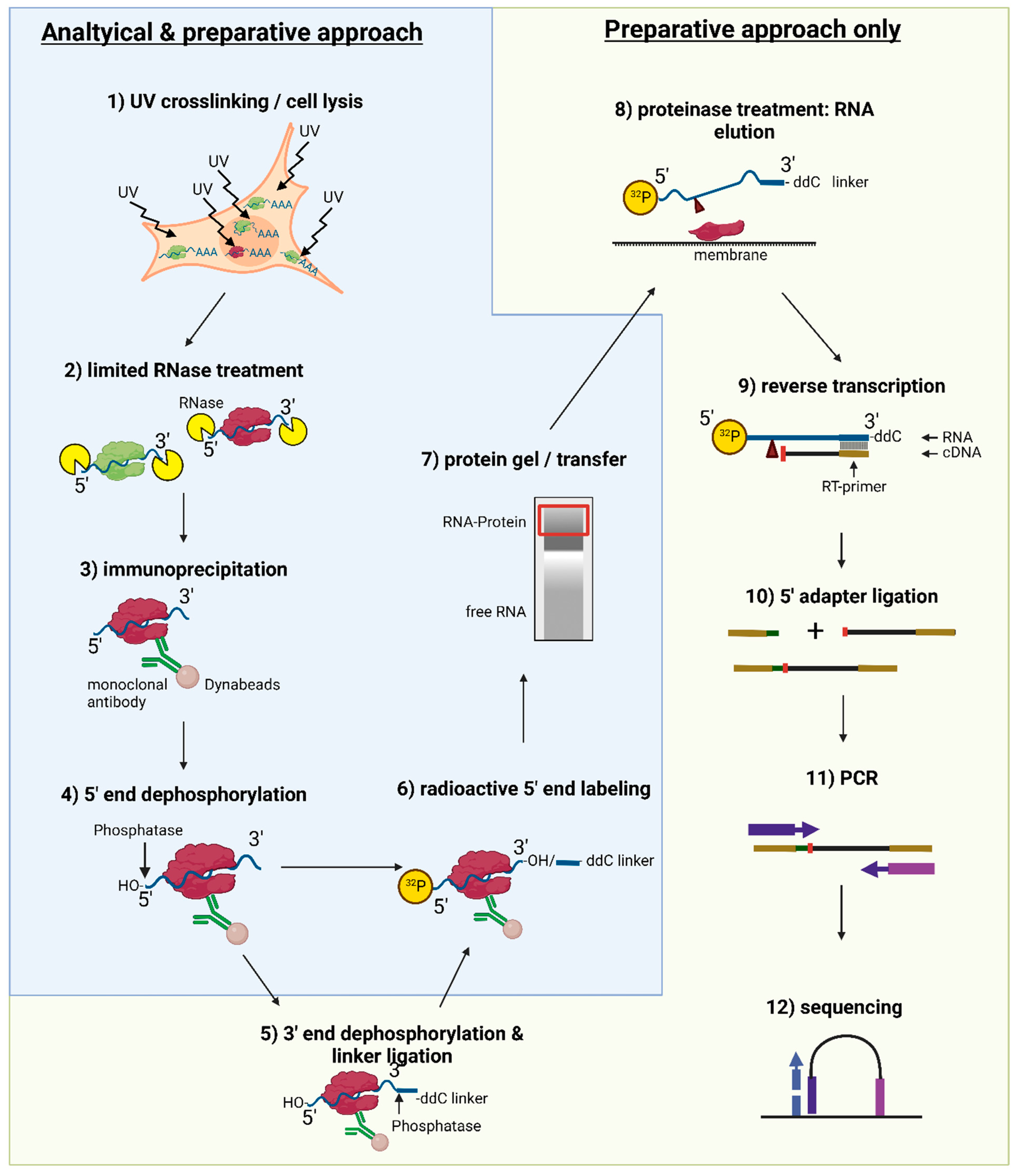
iCLIP - Transcriptome-wide Mapping of Protein-RNA Interactions with Individual Nucleotide Resolution | Protocol (Translated to French)
MPs | Free Full-Text | Studying miRNA–mRNA Interactions: An Optimized CLIP-Protocol for Endogenous Ago2-Protein

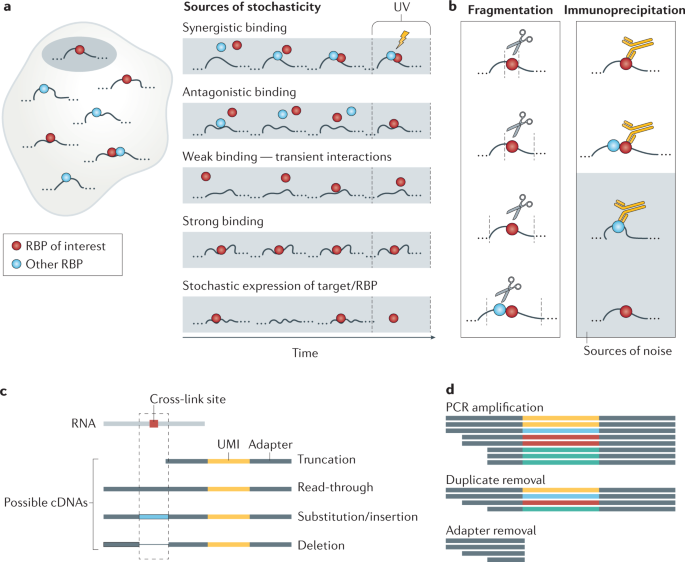
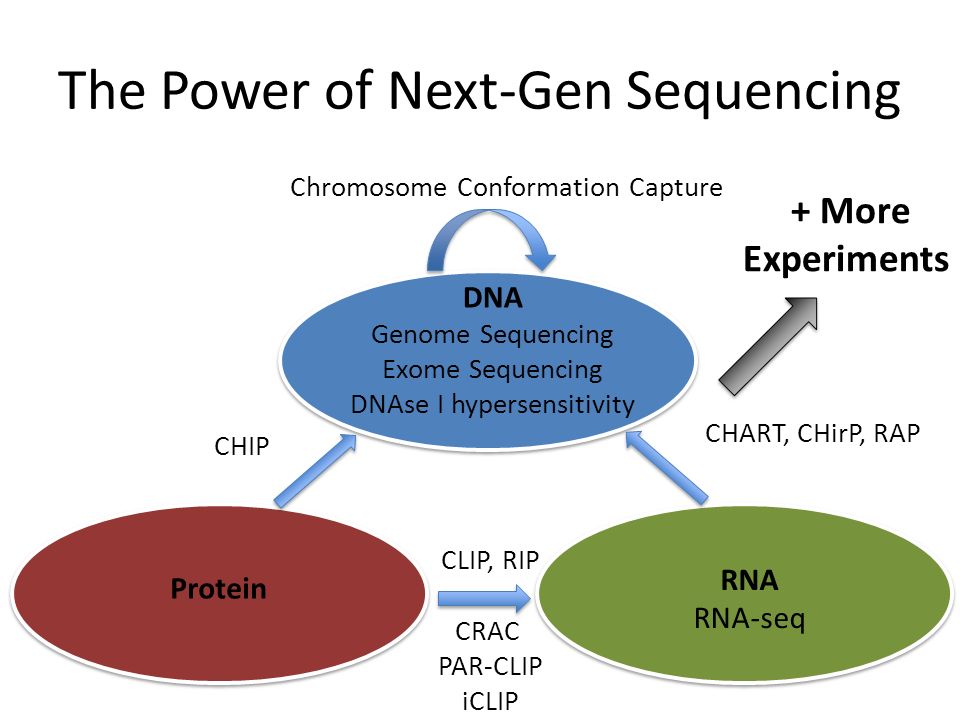
Global Approaches in Studying RNA-Binding Protein Interaction Networks: Trends in Biochemical Sciences

Denaturing CLIP, dCLIP, Pipeline Identifies Discrete RNA Footprints on Chromatin-Associated Proteins and Reveals that CBX7 Targets 3′ UTRs to Regulate mRNA Expression - ScienceDirect

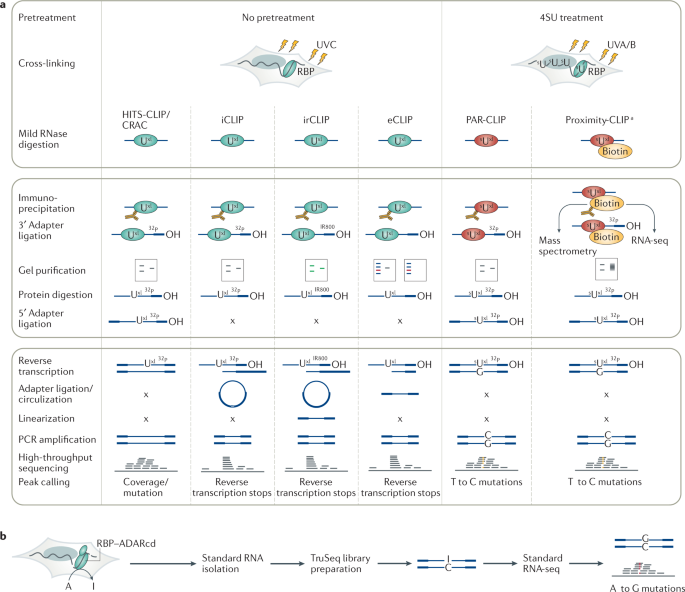
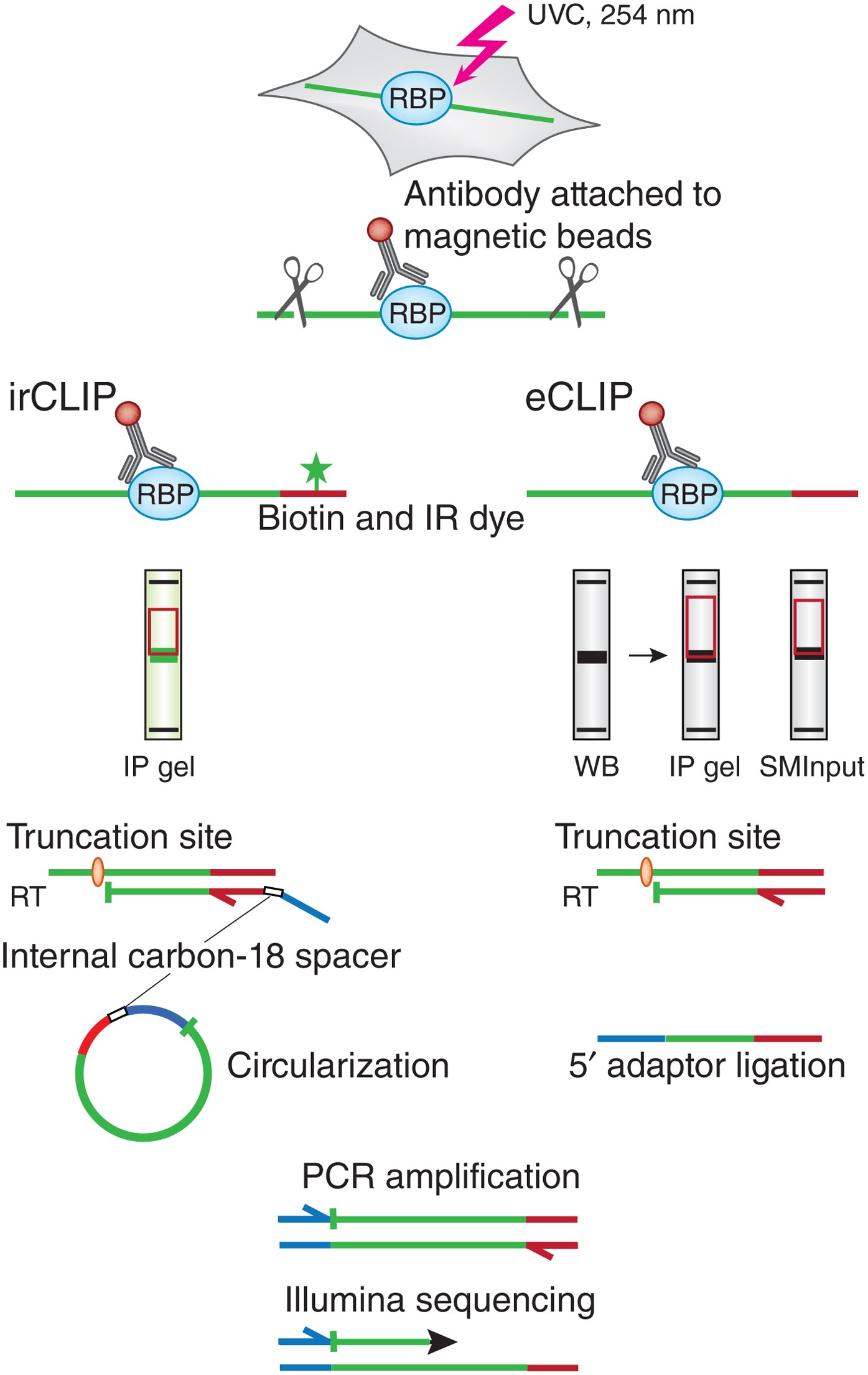


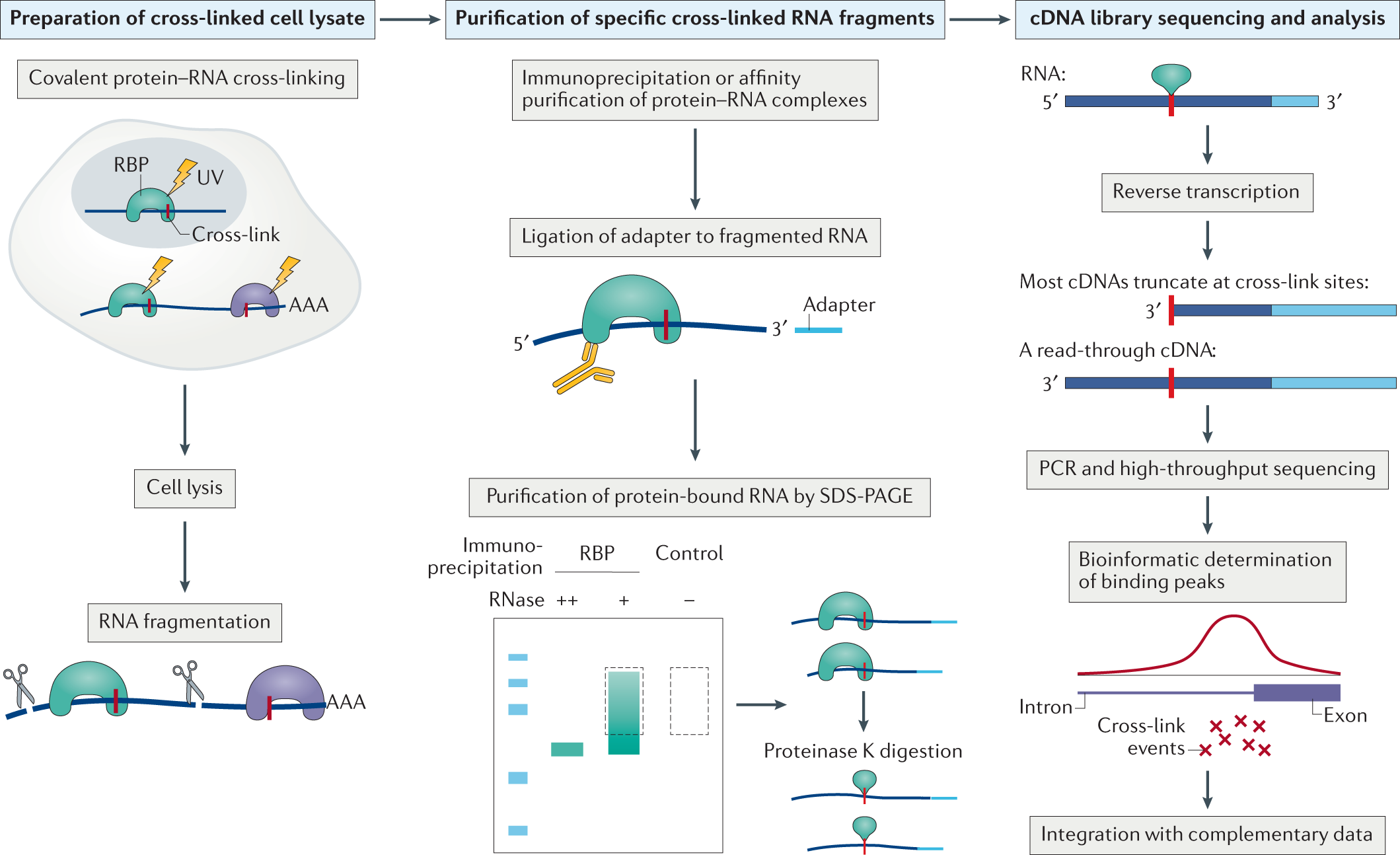
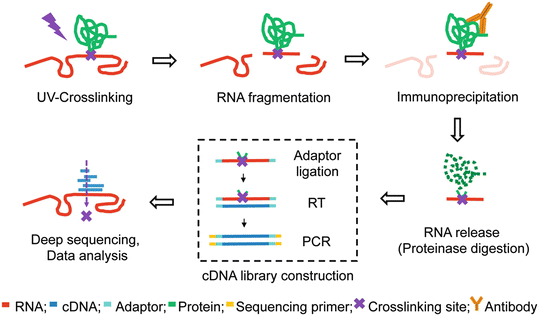
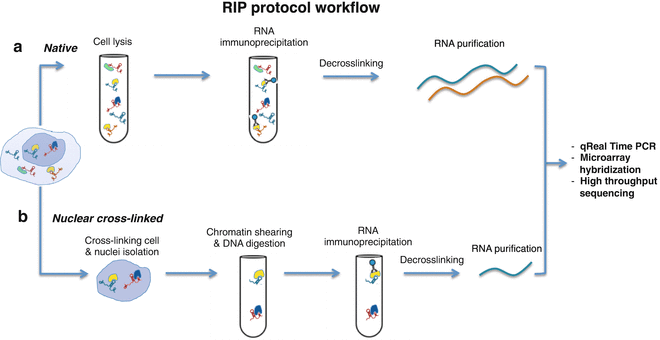
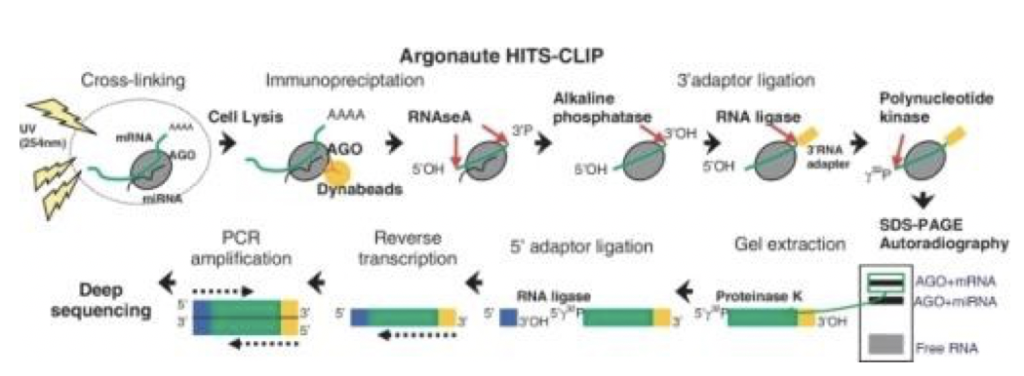
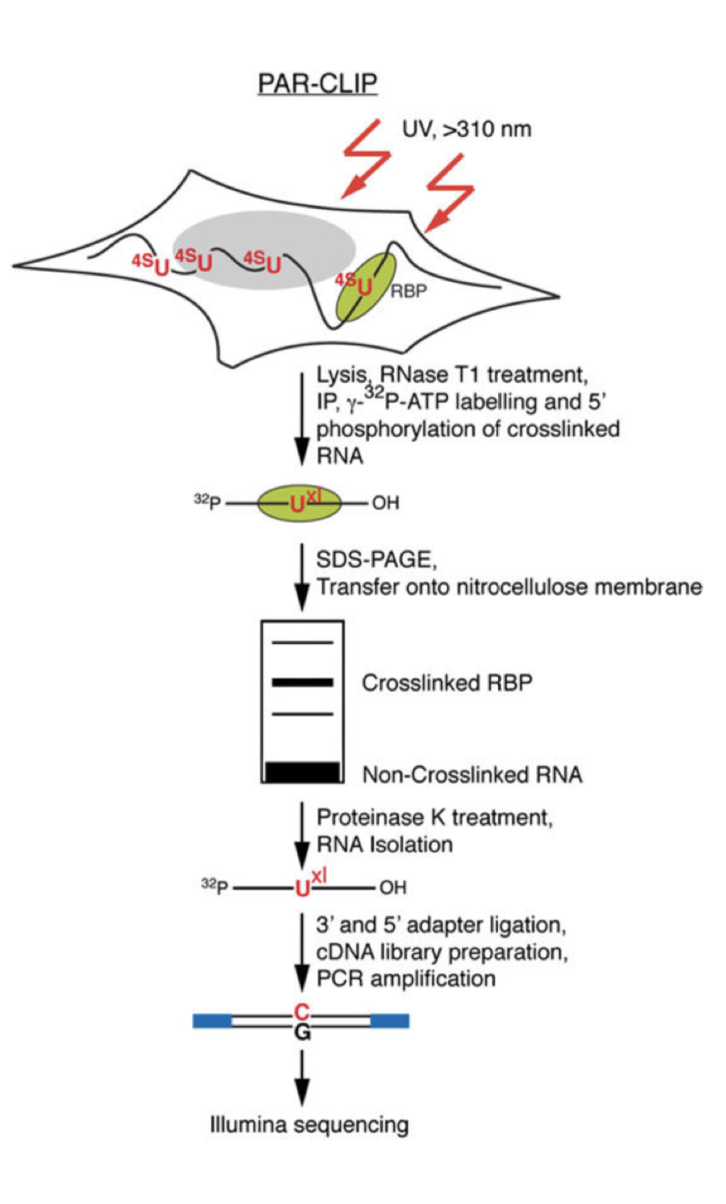
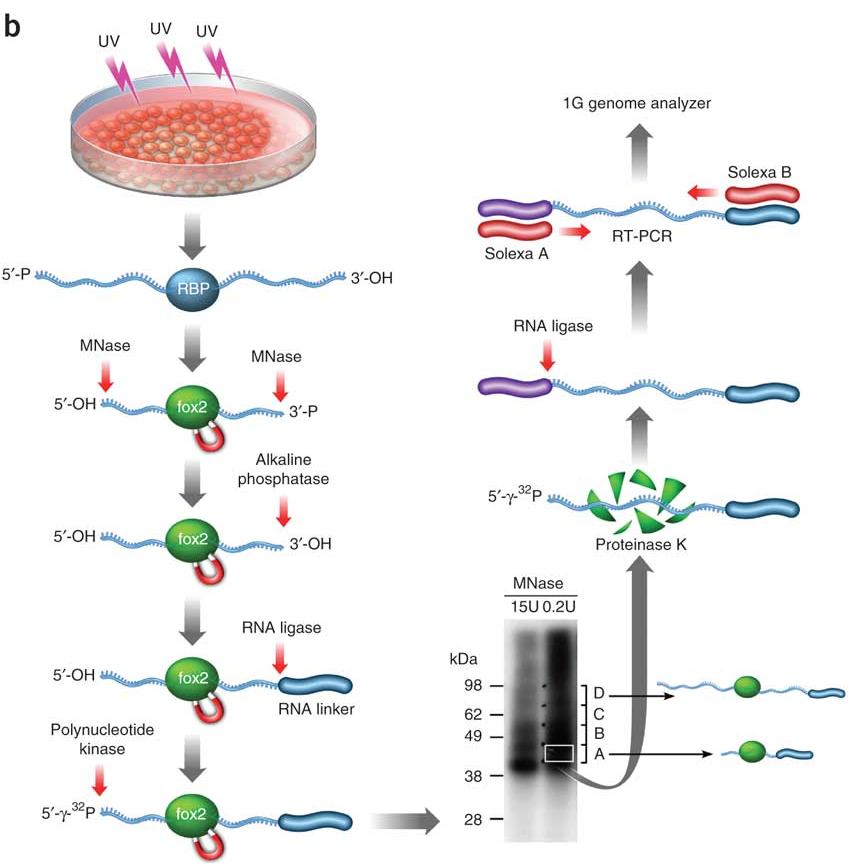
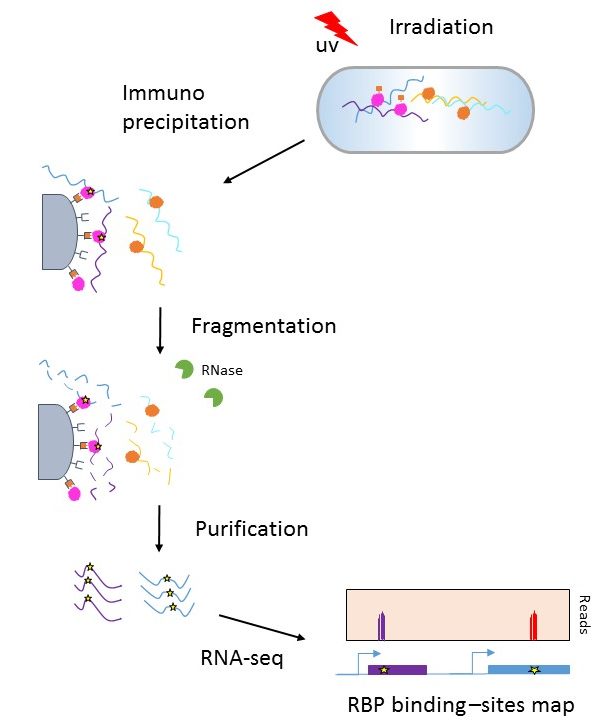

![PDF] Current methods in the analysis of CLIP-Seq data | Semantic Scholar PDF] Current methods in the analysis of CLIP-Seq data | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/e504d75c2c48b19613a796fb54083b71e3781289/2-Figure1-1.png)

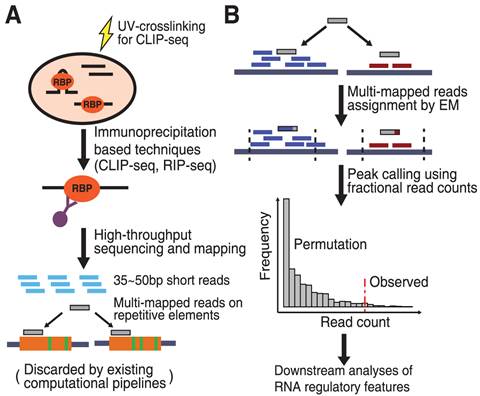
![PDF] CLIPSeqTools—a novel bioinformatics CLIP-seq analysis suite | Semantic Scholar PDF] CLIPSeqTools—a novel bioinformatics CLIP-seq analysis suite | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/6bbd241680ec0f3d6a86dec738a6b864ea5048c3/2-Figure1-1.png)